May 31, 2010
Listening to Mozart does not make you smart
Intelligence doi:10.1016/j.intell.2010.03.001
Mozart effect–Shmozart effect: A meta-analysis
Jakob Pietschnig et al.
The transient enhancement of performance on spatial tasks in standardized tests after exposure to the first movement “allegro con spirito” of the Mozart sonata for two pianos in D major (KV 448) is referred to as the Mozart effect since its first observation by Rauscher, Shaw, and Ky (1993). These findings turned out to be amazingly hard to replicate, thus leading to an abundance of conflicting results. Sixteen years after initial publication we conduct the so far largest, most comprehensive, and up-to-date meta-analysis (nearly 40 studies, over 3000 subjects), including a diversity of unpublished research papers to finally clarify the scientific record about whether or not a specific Mozart effect exists. We could show that the overall estimated effect is small in size (d = 0.37, 95% CI [0.23, 0.52]) for samples exposed to the Mozart sonata KV 448 and samples that had been exposed to a non-musical stimulus or no stimulus at all preceding spatial task performance. Additionally, calculation of effect sizes for samples exposed to any other musical stimulus and samples exposed to a non-musical stimulus or no stimulus at all yielded effects similar in strength (d = 0.38, 95% CI [0.13, 0.63]), whereas there was a negligible effect between the two music conditions (d = 0.15, 95% CI [0.02, 0.28]). Furthermore, formal tests yielded evidence for confounding publication bias, requiring downward correction of effects. The central finding of the present paper however, is certainly the noticeably higher overall effect in studies performed by Rauscher and colleagues than in studies performed by other researchers, indicating systematically moderating effects of lab affiliation. On the whole, there is little evidence left for a specific, performance-enhancing Mozart effect.
Link
May 30, 2010
ESHG 2010 abstracts
P10.31 - Pigmentation gene MC1R shows strong genetic patterning in Eurasia
We present a comprehensive analysis of allele/haplotype frequencies from five functional SNPs (rs1805005, rs2228479, rs1805007, rs1805008, and rs885479) in MC1R throughout Eurasia, including from 12,151 individuals from 141 regional populations, focussing on novel genotype data from 38 Central Asian populations.P10.39 - Genetic variation in Bulgarians: a mitochondrial DNA perspective
The structure and diversity of the Bulgarian mitochondrial DNA (mtDNA) gene pool is still almost unknown. In the present study, we have evaluated the extent and nature of mtDNA variation in the largest Bulgarian sample to date, comprising 855 healthy unrelated subjects from across the country.P10.41 - Mitochondrial genome diversity in Ulchi, the tungusic-speaking tribe of the Russian Far East
The present report is based on the study of mtDNA variation in Ulchi (n=74), a Tungusic-speaking tribe of hunters and fishermen dispersed along the lakes and reaches of the Lower Amur. MtDNA analysis revealed 39 distinct mtDNA haplotypes belonging to 21 Eurasian haplogroups C2-C3, D3-D8, D11, G1-G2, M7-M9, Z, B, F, N9, Y and U4, with overall N macrohaplogroup derivatives frequency 53%, M - 43%, and R - 4%.P10.62 - Genetic structure of Western Caucasus populations on the base of uniparental polymorphisms
We have analyzed 52 markers in coding region of the mtDNA and 48 markers in the non-recombining part of the Y-chromosome in 592 individuals representing five populations from western Caucasus (Abkhazians, Adyghes, Abazins, Georgians, and Circassians). Y-chromosome haplogroups G-M201 and J2 (J-M172) account for more than 50% of all haplogroup diversity in the studied populations. Haplogroup G-M201 in the Western Caucasus populations is represented only by subclade G2a (G-P15) with the insignificantly low exception in the Adyghe population where G1a (G-P20) amounts to less than 1%. In contrast to high frequency of J2 haplogroup J1 exhibit moderate occurrence and vary from 2 to 6 %. Haplogroup R1a (R-SRY10831.2) is also present in all studied populations.
May 29, 2010
Comparison between morphological and genetic data for Egyin Gol Mongolians
The Egyin Gol necropolis is located in the Egyin Gol Valley (northern Mongolia), near the Egyin Gol River, close to its confluence with the Selenge, a main tributary of Lake Baikal (see Fig. 1). This site has been the subject of a French-Mongolian interdisciplinary research project from 1997 to 1999, which allowed the excavation of 84 graves containing skeletal remains of 99 individuals buried from the third century B.C. to the second century A.D. The graves were organized in three main sectors (A, B, and C) that, based on AMS carbon-14 dating of human bones, progressively expanded from south to north (i.e., Sector A is the oldest followed by Sector B and Sector C). The development of Sector C corresponds to the end of the necropolis and may reflect a Turkish influence on the Xiongnu tribe (Keyser-Tracqui et al., 2003).and:
The results showed, however, that individuals buried in sector C represent a specific kin group clearly differentiated from the rest of the necropolis based on nonmetric
data (Table 4), and confirmed by the genetic data. This might be explained on the basis that these individuals are suggested to be of Turkish origin, based on their shared single paternal lineage, unique in the necropolis and affiliated with Turkish populations (Keyser-Tracqui et al., 2003). However, the sector C individuals share the same maternal lineages with individuals buried in sectors A and B, which could explain the global homogeneity of the population as a whole. The particular characteristics of the sample from sector C suggests a possible shift in the population demographics, caused by the emergence of a Turkish component in the Xiongnu population at the end of the necropolis use and at the end of the first steppe empire led by the Xiongnu. The fact that this particular subgroup of the population buried in sector C was detected by nonmetric traits analysis demonstrated that nonmetric traits are an efficient tool when analyzing population microevolution.
 The high frequency in Kazakhs and Yakuts, with a little spillover in both China and eastern Europe is certainly consistent with a Turkic origin of this haplotype.
The high frequency in Kazakhs and Yakuts, with a little spillover in both China and eastern Europe is certainly consistent with a Turkic origin of this haplotype.American Journal of Physical Anthropology doi:10.1002/ajpa.21322
Comparison between morphological and genetic data to estimate biological relationship: The case of the Egyin Gol necropolis (Mongolia)
François-X. Ricaut et al.
Osseous and dental nonmetric (discrete) traits have long been used to assess population variability and affinity in anthropological and archaeological contexts. However, the full extent to which nonmetric traits can reliably be used as a proxy for genetic data when assessing close or familial relationships is currently poorly understood. This study represents the unique opportunity to directly compare genetic and nonmetric data for the same individuals excavated from the Egyin Gol necropolis, Mongolia. These data were analyzed to consider the general efficacy of nonmetric traits for detecting familial groupings in the absence of available genetic data. The results showed that the Egyin Gol population is quite homogenous both metrically and genetically confirming a previous suggestion that the same people occupied the necropolis throughout the five centuries of its existence. Kinship analysis detected the presence of potential family burials in the necropolis. Moreover, individuals buried in one sector of the necropolis were differentiated from other sectors on the basis of nonmetric data. This separation is likely due to an outside Turkish influence in the paternal line, as indicated by the results of Y-chromosome analysis. Affinity matrices based on nonmetric and genetic data were correlated demonstrating the potential of nonmetric traits for detecting relationships in the absence of genetic data. However, the strengths of the correlations were relatively low, cautioning against the use of nonmetric traits when the resolution of the familial relationships is low. Am J Phys Anthropol 2010. © 2010 Wiley-Liss, Inc.
Link
May 28, 2010
Genomic ancestry of ethnic Americans (Wang et al. 2010)
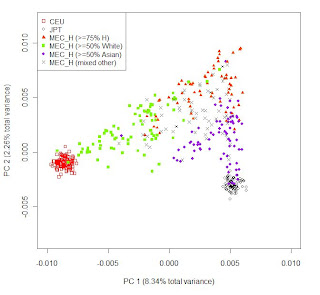 The paper also has quite extensive supplementary material. On the left, part of Figure S6: Clustering of the MEC Hawaiians (MEC_H) using the top 2 PCs from PCA on the HapMap European and East Asian samples plus MEC_H using 2509 SNPs.
The paper also has quite extensive supplementary material. On the left, part of Figure S6: Clustering of the MEC Hawaiians (MEC_H) using the top 2 PCs from PCA on the HapMap European and East Asian samples plus MEC_H using 2509 SNPs. The above (from Figure S2) depicts Japanese Americans on the PC axes determined by all populations. Outliers deviate towards Europeans. In some cases this makes sense, as the individuals report a White parent (1J 1W plus unlabeled dots), but there are a couple dots reporting two Japanese parents that still deviate towards Whites. Assuming no clerical error, this is probably a case of cryptic White ancestry, or, alternatively, a case of of two parents who considered themselves Japanese even though at least one of them had substantial Caucasoid ancestry.
The above (from Figure S2) depicts Japanese Americans on the PC axes determined by all populations. Outliers deviate towards Europeans. In some cases this makes sense, as the individuals report a White parent (1J 1W plus unlabeled dots), but there are a couple dots reporting two Japanese parents that still deviate towards Whites. Assuming no clerical error, this is probably a case of cryptic White ancestry, or, alternatively, a case of of two parents who considered themselves Japanese even though at least one of them had substantial Caucasoid ancestry. Once again, we see a main pink cluster dominated by persons reporting 100% W ancestry, with deviations in two directions: top-right, towards East Asians, and top-left, towards Sub-Saharan Africans. There are clear cases where self-reported ancestry does not match reality, e.g., (i) the group of 4 pink dots on the top right showing clear evidence of East Asian admixture despite reporting 100% W ancestry, or (ii) a set of non-pink dots right in the middle of the pink cluster, which appear (at least in the first two PCs) to be indistinguishable from other Whites, but report mixed ancestry.
Once again, we see a main pink cluster dominated by persons reporting 100% W ancestry, with deviations in two directions: top-right, towards East Asians, and top-left, towards Sub-Saharan Africans. There are clear cases where self-reported ancestry does not match reality, e.g., (i) the group of 4 pink dots on the top right showing clear evidence of East Asian admixture despite reporting 100% W ancestry, or (ii) a set of non-pink dots right in the middle of the pink cluster, which appear (at least in the first two PCs) to be indistinguishable from other Whites, but report mixed ancestry.Human Genetics doi:10.1007/s00439-010-0841-4
Self-reported ethnicity, genetic structure and the impact of population stratification in a multiethnic study
Hansong Wang et al.
It is well-known that population substructure may lead to confounding in case–control association studies. Here, we examined genetic structure in a large racially and ethnically diverse sample consisting of five ethnic groups of the Multiethnic Cohort study (African Americans, Japanese Americans, Latinos, European Americans and Native Hawaiians) using 2,509 SNPs distributed across the genome. Principal component analysis on 6,213 study participants, 18 Native Americans and 11 HapMap III populations revealed four important principal components (PCs): the first two separated Asians, Europeans and Africans, and the third and fourth corresponded to Native American and Native Hawaiian (Polynesian) ancestry, respectively. Individual ethnic composition derived from self-reported parental information matched well to genetic ancestry for Japanese and European Americans. STRUCTURE-estimated individual ancestral proportions for African Americans and Latinos are consistent with previous reports. We quantified the East Asian (mean 27%), European (mean 27%) and Polynesian (mean 46%) ancestral proportions for the first time, to our knowledge, for Native Hawaiians. Simulations based on realistic settings of case–control studies nested in the Multiethnic Cohort found that the effect of population stratification was modest and readily corrected by adjusting for race/ethnicity or by adjusting for top PCs derived from all SNPs or from ancestry informative markers; the power of these approaches was similar when averaged across causal variants simulated based on allele frequencies of the 2,509 genotyped markers. The bias may be large in case-only analysis of gene by gene interactions but it can be corrected by top PCs derived from all SNPs.
Link
Accuracy of molecular dating with the rho statistic questioned (Cox et al. 2008)
I have argued before about the influence on population demography on molecular age estimation methods, as well as the generally large confidence intervals associated with such methods. Quite often papers ignore demography in calculating confidence intervals, but it's not only mutation rate uncertainty, but also population size dynamics, generation length, and other factors that contribute to stretch confidence intervals wide.
In general, there is usually lack of knowledge about demography; some models are patently false, e.g., models of constant population size are incompatible with lineages with multi-million extant descendants. While I believe that in the last few thousand years population growth has been more or less continuous -with short fluctuations that have negligible effects on diversity- things become more complex as we move further into the past: population sizes shrink and stochastic factors take the upper hand.
We've seen time and again that inferences from modern populations don't jibe well with prehistoric DNA samples, so any paper that points out the limitations of such inferences is a welcome addition to the literature.
Human Biology 80(4):335-357. 2008
doi: 10.3378/1534-6617-80.4.335
Accuracy of Molecular Dating with the Rho Statistic: Deviations from Coalescent Expectations Under a Range of Demographic Models
The ρ statistic is commonly used to infer chronological dates for molecular lineages, especially from mitochondrial DNA sequences obtained in anthropological contexts. Since this approach was described 12 years ago, it has been applied to estimate molecular dates in more than 200 studies, including some published in top-tier journals. However, this method has not been well evaluated, and the accuracy of dates obtained from the ρ statistic remains unknown, especially for genetic data collected from populations with complex demographic histories. Here, molecular dates inferred from ρ are compared against coalescent expectations from a range of demographic models. This exercise reveals considerable inaccuracy. Molecular dates based on ρ have a slight downward bias with large asymmetric variance and commonly exhibit substantial type I error rates, where the true age of a lineage falls outside the 95% confidence bounds derived from the variance of ρ. Furthermore, demography proves to be a strong confounding factor in estimating molecular dates accurately, especially for populations in which bottlenecks, founder events, and size changes have played important historical roles. Therefore considerable caution should be applied to inferences made from molecular dates based on the ρ statistic, many of which may be misleading and warrant considerable
May 27, 2010
Origin of domestic chickens (Sawai et al. 2010)
The Origin and Genetic Variation of Domestic Chickens with Special Reference to Junglefowls Gallus g. gallus and G. varius
Hiromi Sawai et al.
It is postulated that chickens (Gallus gallus domesticus) became domesticated from wild junglefowls in Southeast Asia nearly 10,000 years ago. Based on 19 individual samples covering various chicken breeds, red junglefowl (G. g. gallus), and green junglefowl (G. varius), we address the origin of domestic chickens, the relative roles of ancestral polymorphisms and introgression, and the effects of artificial selection on the domestic chicken genome. DNA sequences from 30 introns at 25 nuclear loci are determined for both diploid chromosomes from a majority of samples. The phylogenetic analysis shows that the DNA sequences of chickens, red and green junglefowls formed reciprocally monophyletic clusters. The Markov chain Monte Carlo simulation further reveals that domestic chickens diverged from red junglefowl 58,000±16,000 years ago, well before the archeological dating of domestication, and that their common ancestor in turn diverged from green junglefowl 3.6 million years ago. Several shared haplotypes nonetheless found between green junglefowl and chickens are attributed to recent unidirectional introgression of chickens into green junglefowl. Shared haplotypes are more frequently found between red junglefowl and chickens, which are attributed to both introgression and ancestral polymorphisms. Within each chicken breed, there is an excess of homozygosity, but there is no significant reduction in the nucleotide diversity. Phenotypic modifications of chicken breeds as a result of artificial selection appear to stem from ancestral polymorphisms at a limited number of genetic loci.
Link
May 26, 2010
Genetic Structure of the Spanish Population (Gayan et al. 2010)
Genetic Structure of the Spanish Population
Javier Gayan et al.
Abstract
Background
Genetic admixture is a common caveat for genetic association analysis. Therefore, it is important to characterize the genetic structure of the population under study to control for this kind of potential bias.
Results
In this study we have sampled over 800 unrelated individuals from the population of Spain, and have genotyped them with a genome-wide coverage. We have carried out linkage disequilibrium, haplotype, population structure and copy-number variation (CNV) analyses, and have compared these estimates of the Spanish population with existing data from similar efforts.
Conclusions
In general, the Spanish population is similar to the Western and Northern Europeans, but has a more diverse haplotypic structure. Moreover, the Spanish population is also largely homogeneous within itself, although patterns of micro-structure may be able to predict locations of origin from distant regions. Finally, we also present the first characterization of a CNV map of the Spanish population. These results and original data are made available to the scientific community.
Link
Lathyrus consumption in Late Bronze and Iron Age Israel
Journal of Archaeological Science doi:10.1016/j.jas.2010.05.008
Lathyrus Consumption in Late Bronze and Iron Age Sites in Israel: An Aegean Affinity
Yael Mahler-Slasky et al.
Abstract
This paper presents new evidence, together with previous findings, for the appearance of charred seeds of Lathyrus sativus (grass pea)/L. cicera. This grain legume was a food staple in ancient times, principally in the Aegean region, but also appeared sporadically and in a limited way in the archaeological record of the southern Levant. It is encountered there first in the Late Bronze Age but disappears in the record at the end of the Iron Age. Although a palatable, nutritious plant adapted for growing under adverse conditions, its seeds can be toxic when consumed in large quantities. Apparently L. sativus/cicera made its way to the lowlands of the southern Levant, either by trade or with Philistine immigrants. It is absent at other south Levantine Bronze Age (i.e., Canaanite) and Iron Age sites and it remained a food component in the southern coastal region (i.e., Philistia, the region associated with the biblical Philistines) up to the end of Iron Age II, suggesting a possible ethnic association. Evidence of L. sativus/cicera joins that of another Aegean archaeobotanical import from an earlier, Middle Bronze Age II context, L. clymenum, found at Tel Nami, a coastal site farther to the north of the region.
May 25, 2010
mtDNA of Gelao from Southwest China
 Forensic Sci Int Genet. 2010 May 20. [Epub ahead of print]
Forensic Sci Int Genet. 2010 May 20. [Epub ahead of print]Mitochondrial DNA polymorphisms in Gelao ethnic group residing in Southwest China.
Liu C, Wang SY, Zhao M, Xu ZY, Hu YH, Chen F, Zhang RZ, Gao GF, Yu YS, Kong QP.
Abstract
Gelao ethnic group, an aboriginal population residing in southwest China, has undergone a long and complex evolutionary process. To investigate the genetic structure of this ancient ethnic group, mitochondrial DNA (mtDNA) polymorphisms of 102 Gelao individuals were collected and analyzed in this study. With the aid of the information extracted from control-region hypervariable segments (HVSs) I and II as well as some necessary coding-region segments, phylogenetic status of all mtDNAs under study were determined by means of classifying into various defined haplogroups. The southern-prevalent haplogroups B, R9, and M7 account for 45.1% of the gene pool, whereas northern-prevalent haplogroups A, D, G, N9, and M8 consist of 39.2%. Haplogroup distribution indicates that the Gelao bears signatures of southern populations and possesses some regional characters. In the PC map, Gelao clusters together with populations with Bai-Yue tribe origin as well as the local Han and the Miao. The results demonstrate the complexity of Gelao population and the data can well supplement the China mtDNA database.
Link
May 22, 2010
Origin of Gelong from Hainan island
J Hum Genet. doi:10.1038/jhg.2010.50.
Genetic origin of Kadai-speaking Gelong people on Hainan island viewed from Y chromosomes.
Li D, Sun Y, Lu Y, Mustavich LF, Ou C, Zhou Z, Li S, Jin L, Li H.
The government of China defined 56 official ethnicities for the ethnic groups in China for political purposes; however, there are many more than 56 ethnic groups. Therefore, similar groups must be pooled for registry, and the so-called ethnicity identification is an important official mission in China. Here, we showed how genetics can help in the ethnicity identification for the Gelong people on Hainan Island. The Gelong speak a Kadai language whose other speakers (officially of the Gelao ethnicity) are all far in the southwest of China. Being registered as a Han ethnicity, the Gelong lost all the benefits assigned to the minorities. Y-chromosome typing was performed in a sample of 78 individuals. Twenty single nucleotide polymorphisms (SNPs) and seven short tandem repeats (STRs) were typed and eight haplogroups were detected, among which haplogroup O1a(*) was the most dominant. Compared with the Y haplogroups of the populations in south China, the Gelong were found to be closest to the Gelao and the Hlai using principal components (PCs) analysis, dendrogram clustering and STR networks. The genetic similarity between the Gelong and the Hlai may have resulted from the gene flow during thousands of years of neighboring history on Hainan Island, whereas the similarity between the Gelong and the Gelao may have resulted from their common ancestry because there is less possibility of gene flow over such a far distance. As both linguistic and genetic evidence support the similarity between the Gelong and the Gelao, we suggest that the Gelong register as Gelao for their official ethnicity. However, this identification is invalid until it is accepted by the Gelong people themselves and the Hainan government.
Link
May 21, 2010
Genome-wide structure of West Cape Coloured population (de Wit et al. 2010)

Human Genetics doi:10.1007/s00439-010-0836-1
Genome-wide analysis of the structure of the South African Coloured Population in the Western Cape
Erika de Wit et al.
Admixed populations present unique opportunities to discover the genetic factors underlying many multifactorial diseases. The geographical position and complex history of South Africa has led to the establishment of the unique admixed population known as the South African Coloured. Not much is known about the genetic make-up of this population, and the historical record is patchy. We genotyped 959 individuals from the Western Cape area, self-identified as belonging to this population, using the Affymetrix 500k genotyping platform. This resulted in nearly 75,000 autosomal SNPs that could be compared with populations represented in the International HapMap Project and the Human Genome Diversity Project. Analysis by means of both the admixture and linkage models in STRUCTURE revealed that the major ancestral components of this population are predominantly Khoesan (32–43%), Bantu-speaking Africans (20–36%), European (21–28%) and a smaller Asian contribution (9–11%), depending on the model used. This is consistent with historical data. While of great historical and genealogical interest, this information is also essential for future admixture mapping of disease genes in this population.
May 18, 2010
Soil and climate as economic destiny
 Figure 1 (on the left) Modelled “suitability” (probability of occurrence, Maxent) for (A) agriculture, (B) sedentary animal husbandry, (C) nomadic pastoralism, and (D) hunting and gathering.
Figure 1 (on the left) Modelled “suitability” (probability of occurrence, Maxent) for (A) agriculture, (B) sedentary animal husbandry, (C) nomadic pastoralism, and (D) hunting and gathering.Our simplistic exercise showed that a “geo-deterministic” approach can predict surprisingly many features of human cultural geography without any explicit cultural or historical assumptions. Although many deviations require further factors for a satisfying explanation, the nature of these deviations invite the generation of hypotheses for further research. Our ‘null model’ offers a highly parsimonious, empirically supported explanation to the question of why some regions are more “powerful” than others, supplementing the idea of a historical effect operating through the timing of transition to agriculture [2].I really like this type of paper that looks at a simple explanation for a phenomenon, in this case, economic output as a product of soil/climate quality and suitability. As the authors point out, their "null" model has its limitations, and it is precisely in regions of the world where its predictions do not match the observations, that we should look for additional factors (besides soil and climate) to explain economic traits.
In some instances, our model may actually provide a simpler explanation. Putterman [10], for example, suggested that the dominance of Western European cultures indicates the transmission of “civilization” traits (other than knowledge on agriculture) from regions of first domestication. Our data indicate that higher climatic suitability may have been sufficient for Europe to “catch up”, allowing for much higher population densities than, e.g., the “fertile crescent” region. Models of “suitability” under past climatic scenarios may be helpful to evaluate this. Similarly, models applied to predicted future climatic scenarios may be useful to anticipate changes of the economic suitability of landuse types.
However, our model has clear deviations in some regions that may well be explicable by the availability of animals and plants suitable for domestication (e.g., central Africa). Apart from that, and more importantly, we know that human societies and economies went through historical development, so ignoring history may not always be the best strategy to understand causalities. This problem occurs also with other research questions in biogeography, e.g. when investigating global biodiversity patterns [45]–[47]. Nevertheless, our ‘null model’ will be a useful tool in identifying regions that require further investigation to understand additional processes that shape the distribution and performance of human economic traits.
Related: Soil and Greek temples.
PLoS ONE 5(5): e10416. doi:10.1371/journal.pone.0010416
Is the Spatial Distribution of Mankind's Most Basic Economic Traits Determined by Climate and Soil Alone?
Jan Beck, Andrea Sieber
Abstract
Background
Several authors, most prominently Jared Diamond (1997, Guns, Germs and Steel), have investigated biogeographic determinants of human history and civilization. The timing of the transition to an agricultural lifestyle, associated with steep population growth and consequent societal change, has been suggested to be affected by the availability of suitable organisms for domestication. These factors were shown to quantitatively explain some of the current global inequalities of economy and political power. Here, we advance this approach one step further by looking at climate and soil as sole determining factors.
Methodology/Principal Findings
As a simplistic ‘null model’, we assume that only climate and soil conditions affect the suitability of four basic landuse types – agriculture, sedentary animal husbandry, nomadic pastoralism and hunting-and-gathering. Using ecological niche modelling (ENM), we derive spatial predictions of the suitability for these four landuse traits and apply these to the Old World and Australia. We explore two aspects of the properties of these predictions, conflict potential and population density. In a calculation of overlap of landuse suitability, we map regions of potential conflict between landuse types. Results are congruent with a number of real, present or historical, regions of conflict between ethnic groups associated with different landuse traditions. Furthermore, we found that our model of agricultural suitability explains a considerable portion of population density variability. We mapped residuals from this correlation, finding geographically highly structured deviations that invite further investigation. We also found that ENM of agricultural suitability correlates with a metric of local wealth generation (Gross Domestic Product, Purchasing Power Parity).
Conclusions/Significance
From simplified assumptions on the links between climate, soil and landuse we are able to provide good predictions on complex features of human geography. The spatial distribution of deviations from ENM predictions identifies those regions requiring further investigation of potential explanations. Our findings and methodological approaches may be of applied interest, e.g., in the context of climate change.
Link
May 15, 2010
Neanderthals had ancestral Microcephalin gene
The authors write:
Evans and colleagues proposed that haplogroup D originated from a lineage separated from modern humans for 1.1 million years and introgressed into the human gene pool by 37,000 years ago, probably from a Neanderthal stock [4].This is a great paper in view of the recent controversy about modern human-Neandertal admixture. I have been on the skeptic camp, and this certainly strengthens my African structure thesis.
However, simulation approaches have shown that two key hypotheses of this model, namely positive selection and admixture, can be relaxed as long as Eurasia was settled from an African population that was both subdivided and under expansion [5]. Interestingly, variation in neurocranial geometry have recently suggested significant levels of geographic structure among early modern humans from Africa [6]. In addition, no direct empirical evidence supports a third key component of the model, ie. that Neanderthals carried alleles of the D haplogroup.
There is a broad agreement that the contribution of archaic Homo populations to the modern gene pool, if any, must have been very limited [33], [34]. Different lines of evidence concur to suggest that the dispersal of anatomically modern humans from Africa was accompanied by repeated founder effects [35]–[38]. If these founder effects were drastic, most or all gene genealogies should actually be shallow, and hence the occurrence of ancient splits would imply some degree of introgression from archaic human forms. However, different consequences would be expected if only mild founder effects occurred when anatomically modern humans moved out of Africa. Under these conditions, gene trees would have a strong random component, and a certain fraction thereof, even in the absence of selection, would show two highly divergent major lineages [39]. The likelihood of finding gene genealogies with a very old common ancestor and very differentiated lineages would be even higher if the source African population was subdivided and structured genetically before dispersal, which is what most studies clearly suggest [40]–[43]. These theoretical considerations are actually matched by consistent results in simulation studies [5], [34], [44] and by variation in neurocranial geometry, suggesting significant levels of geographic structure among early modern humans from Africa [6].
The Microcephalin Ancestral Allele in a Neanderthal Individual
Martina Lari et al.
Background
The high frequency (around 0.70 worlwide) and the relatively young age (between 14,000 and 62,000 years) of a derived group of haplotypes, haplogroup D, at the microcephalin (MCPH1) locus led to the proposal that haplogroup D originated in a human lineage that separated from modern humans >1 million years ago, evolved under strong positive selection, and passed into the human gene pool by an episode of admixture circa 37,000 years ago. The geographic distribution of haplogroup D, with marked differences between Africa and Eurasia, suggested that the archaic human form admixing with anatomically modern humans might have been Neanderthal.
Methodology/Principal Findings
Here we report the first PCR amplification and high- throughput sequencing of nuclear DNA at the microcephalin (MCPH1) locus from Neanderthal individual from Mezzena Rockshelter (Monti Lessini, Italy). We show that a well-preserved Neanderthal fossil dated at approximately 50,000 years B.P., was homozygous for the ancestral, non-D, allele. The high yield of Neanderthal mtDNA sequences of the studied specimen, the pattern of nucleotide misincorporation among sequences consistent with post-mortem DNA damage and an accurate control of the MCPH1 alleles in all personnel that manipulated the sample, make it extremely unlikely that this result might reflect modern DNA contamination.
Conclusions/Significance
The MCPH1 genotype of the Monti Lessini (MLS) Neanderthal does not prove that there was no interbreeding between anatomically archaic and modern humans in Europe, but certainly shows that speculations on a possible Neanderthal origin of what is now the most common MCPH1 haplogroup are not supported by empirical evidence from ancient DNA.
Link
May 14, 2010
The Mediterranean as barrier to gene flow (Athanasiadis et al. 2010)
BMC Evolutionary Biology 2010, 10:84doi:10.1186/1471-2148-10-84
The Mediterranean Sea as a barrier to gene flow: evidence from variation in and around the F7 and F12 genomic regions
Georgios Athanasiadis et al.
Abstract
Background
The Mediterranean has a long history of interactions among different peoples. In this study, we investigate the genetic relationships among thirteen population samples from the broader Mediterranean region together with three other groups from the Ivory Coast and Bolivia with a particular focus on the genetic structure between North Africa and South Europe. Analyses were carried out on a diverse set of neutral and functional polymorphisms located in and around the coagulation factor VII and XII genomic regions (F7 and F12).
Results
Principal component analysis revealed a significant clustering of the Mediterranean samples into North African and South European groups consistent with the results from the hierarchical AMOVA, which showed a low but significant differentiation between groups from the two shores. For the same range of geographic distances, populations from each side of the Mediterranean were found to differ genetically more than populations within the same side. To further investigate this differentiation, we carried out haplotype analyses, which provided partial evidence that sub-Saharan gene flow was higher towards North Africa than South Europe.
Conclusions
As there is no consensus between the two genomic regions regarding gene flow through the Sahara, it is hard to reach a solid conclusion about its role in the differentiation between the two Mediterranean shores and more data are necessary to reach a definite conclusion. However our data suggest that the Mediterranean Sea was at least partially a barrier to gene flow between the two shores.
Link
May 13, 2010
mtDNA and trans-Saharan slave trade (Harich et al. 2010)
 The main trans-Saharan slave routes are shown in Figure 1 (on the left). The paper also contains quite useful interpolation maps of the main Sub-Saharan African mtDNA haplogroups, who should be useful for future reference.
The main trans-Saharan slave routes are shown in Figure 1 (on the left). The paper also contains quite useful interpolation maps of the main Sub-Saharan African mtDNA haplogroups, who should be useful for future reference.BMC Evolutionary Biology 2010, 10:138 doi:10.1186/1471-2148-10-138
Background
A proportion of 1/4 to 1/2 of North African female pool is made of typical sub-Saharan lineages, in higher frequencies as geographic proximity to sub-Saharan Africa increases. The Sahara was a strong geographical barrier against gene flow, at least since 5,000 years ago, when desertification affected a larger region, but the Arab trans-Saharan slave trade could have facilitate enormously this migration of lineages. Till now, the genetic consequences of these forced trans-Saharan movements of people have not been ascertained.
Results
The distribution of the main L haplogroups in North Africa clearly reflects the known trans-Saharan slave routes: West is dominated by L1b, L2b, L2c, L2d, L3b and L3d; the Center by L3e and some L3f and L3w; the East by L0a, L3h, L3i, L3x and, in common with the Center, L3f and L3w; while, L2a is almost everywhere. Ages for the haplogroups observed in both sides of the Saharan desert testify the recent origin (holocenic) of these haplogroups in sub-Saharan Africa, claiming a recent introduction in North Africa, further strengthened by the no detection of local expansions.
Conclusions
The interpolation analyses and complete sequencing of present mtDNA sub-Saharan lineages observed in North Africa support the genetic impact of recent trans-Saharan migrations, namely the slave trade initiated by the Arab conquest of North Africa in the seventh century. Sub-Saharan people did not leave traces in the North African maternal gene pool for the time of its settlement, some 40,000 years ago.
Link
May 12, 2010
mtDNA of Tatars from Volga-Ural region (Malyarchuk et al. 2010)
Molecular Biology and Evolution, doi:10.1093/molbev/msq065
Mitogenomic diversity in Tatars from the Volga-Ural region of Russia
B. Malyarchuk et al.
To investigate diversity of mitochondrial gene pool of Tatars inhabiting the territory of the middle Volga River basin, 197 individuals from two populations representing Kazan Tatars and Mishars were subjected for analysis of mitochondrial DNA (mtDNA) control region variation. In addition, 73 mitochondrial genomes of individuals from Mishar population were sequenced completely. It was found that mitochondrial gene pool of the Volga Tatars consists of two parts, but western Eurasian component prevails considerably (84% on average) over eastern Asian one (16%). Eastern Asian mtDNAs detected in Tatars belonged to a heterogeneous set of haplogroups (A, C, D, G, M7, M10, N9a, Y, Z), although only haplogroups A and D were revealed simultaneously in both populations. Complete mtDNA variation study revealed that the age of western Eurasian haplogroups (such as U4, HV0a and H) is less than 18,000 years, thus suggesting re-expansion of Eastern Europeans soon after the Last Glacial Maximum.
Link
May 11, 2010
Complex picture of R-M269 dispersal
Increased Resolution Within Y-Chromosome Haplogroup R1b M269 Sheds Light On The Neolithic Transition In Europe
Early studies on classical polymorphisms have largely been vindicated by the growing tome of information on the genetic structure of European populations, with mtDNA, Y-Chromosome and autosomal markers all combining to give a fundamental pattern of migration from the East. The processes behind this pattern are however, less clear, particularly with regard to uniparental markers. Much debate still rages about how best to use Y and mtDNA to date particular historical movements, or indeed if it is appropriate at all. For example, whilst some progress has been made recently in calibrating the mtDNA clock, the selection of a mutation rate with which to date the Y-Chromosome is contentious, as the two most favoured values can give dates that differ by a factor of three. In order to address this we have investigated the sub-lineages of the common European haplogroup R1b-M269. This haplogroup has been shown to be clinal in Europe, and more recently has been posited to be the result of the Neolithic expansion from the Near East. Here, we use newly characterised SNPs downstream of M269 to produce a refined picture of the haplogroup in Europe, and further show that the diversity of this lineage cannot be entirely attributed to Neolithic migration out of Anatolia. We use simple coalescent simulations to estimate an absolute lower bound for the age of the sub-haplogoups. Rather than originating with the farmers from the East, we suggest that the sub-structure of R1b-M269 visible in Europe today, and thus the great majority of European paternal ancestry, is the result of the interaction between the Neolithic wave of expansion and populations of early Europeans already present in the path of the wave.
Direction of gene flow (to or from Neanderthals, or ...?)
- The uniformity of alleged Neandertal admixture outside Africa is suspect, given that Neandertals were a West Eurasian species
- The observed pattern of Neandertals being closer to non-Africans than to Africans can be well explained by structure in Africa itself, a point which the authors concede (as their Scenario 4), but which has received zero play in the media which -as usual- have jumped on the more easily digestible ("caveman sex") explanation.
Our data on neighbors and variability is unsupportive of the strict forms of a single-origin model but does not conflict with another approach, the model of ‘‘isolation by distance,’’ which predicts that genetic and phenotypic dissimilarity increases with geographic distance (24, 29–31). The metapopulation framework would predict the same because frequency and magnitude of genetic exchange would follow the likelihood of 2 populations to meet, which declines with geographical distance from the early AMH epicenter in Africa. Our fossil AMH data, however, suggest that before there was isolation by distance from Africa, there already existed (at least temporally) isolation by distance within Africa during the Pleistocene.I won't present the reasoning behind these conclusions (the paper is open access anyway), but they have an important implication: seemingly "ancient" contributions to modern humans need not have been acquired by either Neandertals or other archaic (pre-sapiens) populations, but they could also have been acquired by admixture with different branches of anatomically modern humans.
Seemingly ancient contributions to the modern human gene pool (36) have been explained by admixture with archaic forms of Homo, e.g., Neanderthals. Although we cannot rule out such admixture (37), the clear morphological distinction between
AMH and archaic forms of Homo in the light of the proposed ancestral population structure of early AMH to us suggests another underestimated possibility: the genetic exchange between subdivided populations of early AMH as a potential source for ‘‘ancient’’ contributions to the modern human gene pool (9, 36).
May 10, 2010
Origin and dispersal of Y chromosome haplogroup C (Zhong et al. 2010)
Hg C is prevalent in various geographical areas (Figures 1 and 2), including Australia (65.74%), Polynesia (40.52%), Heilongjiang of northeastern China (Manchu, 44.00%), Inner Mongolia (Mongolian, 52.17%; Oroqen, 61.29%), Xinjiang of northwestern China (Hazak, 75.47%), Outer Mongolia (52.80%) and northeastern Siberia (37.41%). Hg C is also present in other regions, extending longitudinally from Sardinia in Southern Europe all the way to Northern Colombia, and latitudinally from Yakutia of Northern Siberia and Alaska of Northern America to India, Indonesia and Polynesia, but absent in Africa.On the structure of haplogroup C:
As shown in Figure 1, most of the subhaplogroups of Hg C have a geographically pronounced distribution. Hg C6, which is defined by a recently identified marker, was not detected in our samples. Hg C1 and C4 are completely restricted to Japan and Australia, respectively, and not detected in the other samples from East Asia and Southeast Asia. Hg C5 occurs in India and its neighboring regions Pakistan and Nepal. In mainland East Asia, four Hg C5 individuals were detected, including two in Xibe, one in Uygur and one in Shanxi Han. Although the dispersal of Hg C2 is relatively wide, its distribution remains limited to Oceania and its neighboring regions, except Australia. In our samples, only three Hg C2 individuals were observed in Eastern Indonesia, which is consistent with previous reports. Hg C3 is the most widespread subhaplogroup, which was detected in Central Asia, South Asia, Southeast Asia, East Asia, Siberia and the Americas, but absent in Oceania. Different subhaplogroups of Hg C that do not overlap between the regions suggest that these individuals have undergone long-time isolation. As these subhaplogroups have a common origin by sharing the M130-derived allele, their geographical distributions enable us to infer the prehistoric migration routes of this lineage.The MDS plot is quite instructive. Notice the duality of Japanese C chromosomes, which parallels what we know about the dual origins of the Japanese. It would be instructive to test European haplogroup C outliers to see where they fall within haplogroup C diversity.
Journal of Human Genetics doi: 10.1038/jhg.2010.40
Global distribution of Y-chromosome haplogroup C reveals the prehistoric migration routes of African exodus and early settlement in East Asia
Hua Zhong et al.
The regional distribution of an ancient Y-chromosome haplogroup C-M130 (Hg C) in Asia provides an ideal tool of dissecting prehistoric migration events. We identified 465 Hg C individuals out of 4284 males from 140 East and Southeast Asian populations. We genotyped these Hg C individuals using 12 Y-chromosome biallelic markers and 8 commonly used Y-short tandem repeats (Y-STRs), and performed phylogeographic analysis in combination with the published data. The results show that most of the Hg C subhaplogroups have distinct geographical distribution and have undergone long-time isolation, although Hg C individuals are distributed widely across Eurasia. Furthermore, a general south-to-north and east-to-west cline of Y-STR diversity is observed with the highest diversity in Southeast Asia. The phylogeographic distribution pattern of Hg C supports a single coastal ‘Out-of-Africa’ route by way of the Indian subcontinent, which eventually led to the early settlement of modern humans in mainland Southeast Asia. The northward expansion of Hg C in East Asia started ~40 thousand of years ago (KYA) along the coastline of mainland China and reached Siberia ~15 KYA and finally made its way to the Americas.
Link
May 07, 2010
Tales of Neanderthal admixture in modern Eurasians
PS: The only potential argument in favor of the authors' hypothesis is the following:
Non-Africans haplotypes match Neandertals unexpectedly often. An alternative approach to detect gene flow from Neandertals into modern humans is to focus on patterns of variation in present-day humans—blinded to information from the Neandertal genome—in order to identify regions that are the strongest candidates for being derived from Neandertals. If these candidate regions match the Neandertals at a higher rate than is expected by chance, this provides additional evidence for gene flow from Neandertals into modern humans.
We thus identified regions in which there is considerably more diversity outside Africa than inside Africa, as might be expected in regions that have experienced gene flow from Neandertals to non-Africans. We used 1,263,750 Perlegen Class A SNPs, identified in individuals of diverse ancestry (78), and found 13 candidate regions of Neandertal ancestry (SOM Text 17). A prediction of Neandertal-to-modern human gene flow is that DNA sequences that entered the human gene pool from Neandertals will tend to match Neandertal more often than their frequency in the present-day human population. To test this prediction, we identified 166 "tag SNPs" that separate 12 of the haplotype clades in non-Africans (OOA) from the cosmopolitan haplotype clades shared between Africans and non-Africans (COS) and for which we had data from the Neandertals. Overall, the Neandertals match the deep clade unique to non- Africans at 133 of the 166 tag SNPs, and 10 of the 12 regions where tag SNPs occur show an excess of OOA over COS sites. Given that the OOA alleles occur at a frequency of much less than 50% in non-Africans (average of 13%, and all less than 30%) (Table 5), the fact that the candidate regions match the Neandertals in 10 of 12 cases (P = 0.019) suggests that they largely derive from Neandertals. The proportion of matches is also larger than can be explained by contamination, even if all Neandertal data were composed of present-day non-African DNA (P = 0.0025) (SOM Text 17).The problem with the above analysis is in the underlined portion. The deep clade is rather unique to non-Americans of African ancestry, as per reference (78), i.e., a very limited sample of Africans. In short, there is no reason to believe that the identified deep clades matching the Neandertals are really unique to non-Africans, and the pattern can be easily explained by geographical structure within Africa itself.
Although gene flow from Neandertals into modern humans when they first left sub-Saharan Africa seems to be the most parsimonious model compatible with the current data, other scenarios are also possible. For example, we cannot currently rule out a scenario in which the ancestral population of present-day non-Africans was more closely related to Neandertals than the ancestral population of present-day Africans due to ancient substructure within Africa (Fig. 6). If after the divergence of Neandertals there was incomplete genetic homogenization between what were to become the ancestors of non-Africans and Africans, present-day non-Africans would be more closely related to Neandertals than are Africans. In fact, old population substructure in Africa has been suggested based on genetic (81) as well as paleontological data (86).
Whether their model is more parsimonious than Scenario 4 is up to the reader. We know that there is population structure in Africa today, and as the authors note there are reasons why this ought to have been true in the past. So, while Scenario 4 makes up a reasonable xtra assumption (the one I describe in my post), the authors' favored scenario does not explain easily how a species that inhabited Western Eurasia (Neandertals) ended up contributing not-too different amounts of DNA to Europeans and Chinese. So, for the time being, I'm sticking to my guns and saying that the paper has uncovered something important, but probably not Neandertal admixture.
Vol. 328. no. 5979, pp. 710 - 722
A Draft Sequence of the Neandertal Genome
Richard E. Green et al.
Neandertals, the closest evolutionary relatives of present-day humans, lived in large parts of Europe and western Asia before disappearing 30,000 years ago. We present a draft sequence of the Neandertal genome composed of more than 4 billion nucleotides from three individuals. Comparisons of the Neandertal genome to the genomes of five present-day humans from different parts of the world identify a number of genomic regions that may have been affected by positive selection in ancestral modern humans, including genes involved in metabolism and in cognitive and skeletal development. We show that Neandertals shared more genetic variants with present-day humans in Eurasia than with present-day humans in sub-Saharan Africa, suggesting that gene flow from Neandertals into the ancestors of non-Africans occurred before the divergence of Eurasian groups from each other.
Link
May 06, 2010
Population structure in Hispanics (Bryc et al. 2010)
The study, published in the May 3 online issue of the Proceedings of the National Academy of Sciences, tested the genetic makeup of 100 individuals of Hispanic/Latino background in the New York tri-state area, including Dominicans, Columbians and Ecuadorians, as well as Mexicans and Puerto Ricans, the two largest Hispanic/Latino ethnic groups in the United States. Currently, Hispanic/Latino Americans comprise 15.4% of the total United States population, or 46.9 million people, and account for the largest ethnic minority in the United States.
"It is important to quantify the relative contributions of ancestry in relation to disease outcome in the Hispanic/Latino population," says study co-author Christopher Velez, a medical student at NYU School of Medicine. "This ethnically appropriate genetic research will be critical to the understanding of disease onset and severity in the United States and in Latin America. It will allow for the development of appropriate genetic tests for this population."
Through their analysis of the entire genome, the researchers found evidence of a significant sex bias consistent with the disproportionate contribution of European male and Native American female ancestry to present day populations. The scientists also found that the patterns of genes in the Hispanic/Latino populations were impacted by proximity to the African slave trade. In fact, Puerto Ricans, Dominicans and Columbians from the Caribbean coast had higher proportions of African ancestry, while Mexicans and Ecuadorians showed the lowest level of African ancestry and the highest Native American ancestry.
European migrant contributors were mostly from the Iberian Peninsula and Southern Europe. Evidence was also found for Middle Eastern and North African ancestry, reflecting the Moorish and Jewish (as well as European) origins of the Iberian populations at the time of colonization of the New World. The Native Americans that most influenced the Hispanic/Latino populations were primarily from local indigenous populations.
Genome-wide patterns of population structure and admixture among Hispanic/Latino populations
Katarzyna Bryc et al.
Abstract
Hispanic/Latino populations possess a complex genetic structure that reflects recent admixture among and potentially ancient substructure within Native American, European, and West African source populations. Here, we quantify genome-wide patterns of SNP and haplotype variation among 100 individuals with ancestry from Ecuador, Colombia, Puerto Rico, and the Dominican Republic genotyped on the Illumina 610-Quad arrays and 112 Mexicans genotyped on Affymetrix 500K platform. Intersecting these data with previously collected high-density SNP data from 4,305 individuals, we use principal component analysis and clustering methods FRAPPE and STRUCTURE to investigate genome-wide patterns of African, European, and Native American population structure within and among Hispanic/Latino populations. Comparing autosomal, X and Y chromosome, and mtDNA variation, we find evidence of a significant sex bias in admixture proportions consistent with disproportionate contribution of European male and Native American female ancestry to present-day populations. We also find that patterns of linkage-disequilibria in admixed Hispanic/Latino populations are largely affected by the admixture dynamics of the populations, with faster decay of LD in populations of higher African ancestry. Finally, using the locus-specific ancestry inference method LAMP, we reconstruct fine-scale chromosomal patterns of admixture. We document moderate power to differentiate among potential subcontinental source populations within the Native American, European, and African segments of the admixed Hispanic/Latino genomes. Our results suggest future genome-wide association scans in Hispanic/Latino populations may require correction for local genomic ancestry at a subcontinental scale when associating differences in the genome with disease risk, progression, and drug efficacy, as well as for admixture mapping.
Link
Portraits of ethnic groups of China

A great series of portraits of Chinese ethnic groups. I think this may be a great resource when reading papers on Chinese genetics and anthropology, as it provides a direct visual snapshot of the groups involved, most of which are probably not familiar to Western readers.





























































